Morizono Laboratory
About the Lab
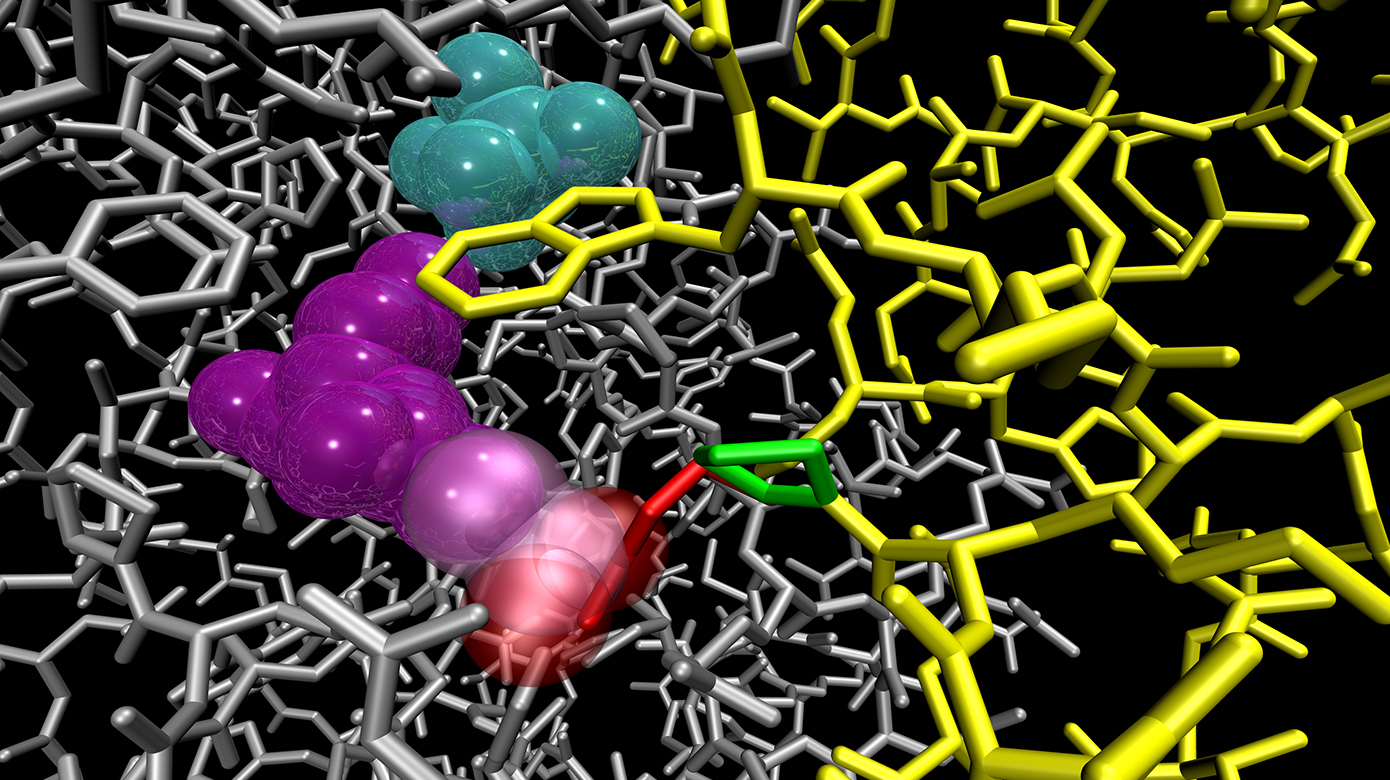
Research in the Morizono laboratory focuses on applications of high-performance informatic and computational methods to visualize and understand human health and disease across a range of biomedical disciplines. This includes bioinformatic analysis of gene regulation, modeling protein structure and stability as a function of mutation, creation of databases incorporating the electronic health record and implementing tools to perform data analysis at large scales. In this way, we create connections between genomics, biochemistry, physiology and clinical applications.
-
Lab Focus Areas
Protein structure function relationships Genomics Biomedical informatics
-
Contact
Hiroki Morizono, PhD Principal Investigator [email protected] 240-531-6584
Featured Publications
-
Criticality: A new concept of severity of illness for hospitalized children
Rivera EAT, Patel AK, Chamberlain JM, Workman TE, Heneghan JA, Redd D, Morizono H, Kim D, Bost JE, Pollack MM PMID: 32932406 Pediatr Crit Care Med doi: 10.1097/PCC.0000000000002560 September (2020) -
A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency
Wang L, Yang Y, Breton C, Bell P, Li M, Zhang J, Che Y, Saveliev A, He Z, White J, Latshaw C, Xu C, McMenamin D, Yu H, Morizono H, Batshaw ML, Wilson JM PMID: 32095520 Sci Adv 6(7):eaax5701. doi: 10.1126/sciadv.aax5701. eCollection February (2020) -
Baseline human gut microbiota profile in healthy people and standard reporting template
King CH, Desai H, Sylvetsky AC, LoTempio J, Ayanyan S, Carrie J, Crandall KA, Fochtman BC, Gasparyan L, Gulzar N, Howell P, Issa N, Krampis K, Mishra L, Morizono H, Pisegna JR, Rao S, Ren Y, Simonyan V, Smith K, VedBrat S, Yao MD, Mazumder R PMID: 31509535 PLoS One 14(9):e0206484. doi: 10.1371/journal.pone.0206484. eCollection September (2019) -
Disease-causing mutations in the promoter and enhancer of the ornithine transcarbamylase gene
Jang YJ, LaBella AL, Feeney TP, Braverman N, Tuchman M, Morizono H, Ah Mew N, Caldovic L PMID: 29282796 Hum Mutat 39(4):527-536. doi: 10.1002/humu.23394. Epub April (2018) -
Enabling precision medicine via standard communication of HTS provenance, analysis, and results
Alterovitz G, Dean D, Goble C, Crusoe MR, Soiland-Reyes S, Bell A, Hayes A, Suresh A, Purkayastha A, King CH, Taylor D, Johanson E, Thompson EE, Donaldson E, Morizono H, Tsang H, Vora JK, Goecks J, Yao J, Almeida JS, Keeney J, Addepalli K, Krampis K, Smith KM, Guo L, Walderhaug M, Schito M, Ezewudo M, Guimera N, Walsh P, Kahsay R, Gottipati S, Rodwell TC, Bloom T, Lai Y, Simonyan V, Mazumder R PMID: 30596645 PLoS Biol 16(12):e3000099. doi: 10.1371/journal.pbio.3000099. eCollection December (2018)
Related News
-

Researchers advance new drug development for fetal alcohol spectrum disorders
November 07, 2025 -
Understanding gut bacteria: forces for good (and sometimes evil)
September 11, 2019 -
Exploring accurate data use that supports clinical judgment
August 07, 2019
